We are sharing MATLAB scripts for data-driven optimization of variable flip angles in three types of MP-GRE sequences often used for T1ρ mapping. Optimized sequences have better accuracy, greater precision, and higher speed—advantages that can add up to T1ρ maps with significantly higher resolution.
For our latest resource for optimizing T1ρ acquisitions, see Learned MR Pulse Sequences for T1ρ Mapping.
The types of sequences that can be optimized with our scripts are:
- magnetization-prepared angle-modulated partitioned k-space spoiled GRE snapshots (MAPSS)
- tailored variable flip angle MP-GRE with magnetization reset (MP-GRE-WR)
- standard magnetization-prepared gradient echo (MP-GRE)
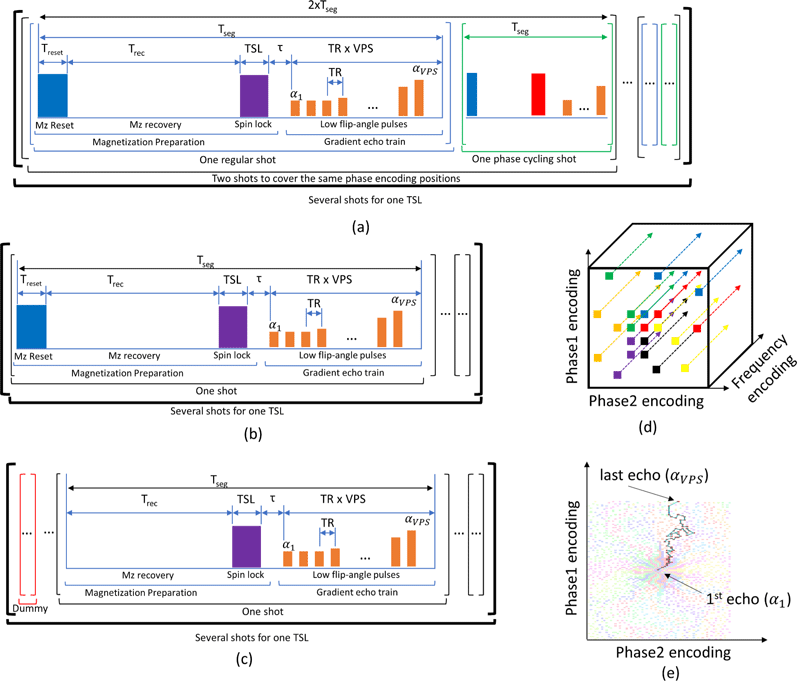
The scripts offer three major improvements:
- better accuracy through adjustment of flip angles to minimize every modeled pulse sequence imperfection
- greater precision through automatically increasing flip angles to improve the sequence’s signal-to-noise ratio while reducing undesirable effects, such as k-space filtering
- higher speed through the use of sequence parameters to capture more data per unit of time (less idle time)
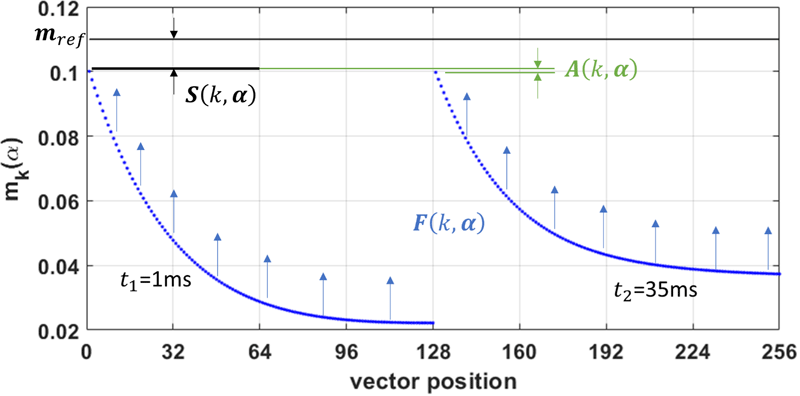
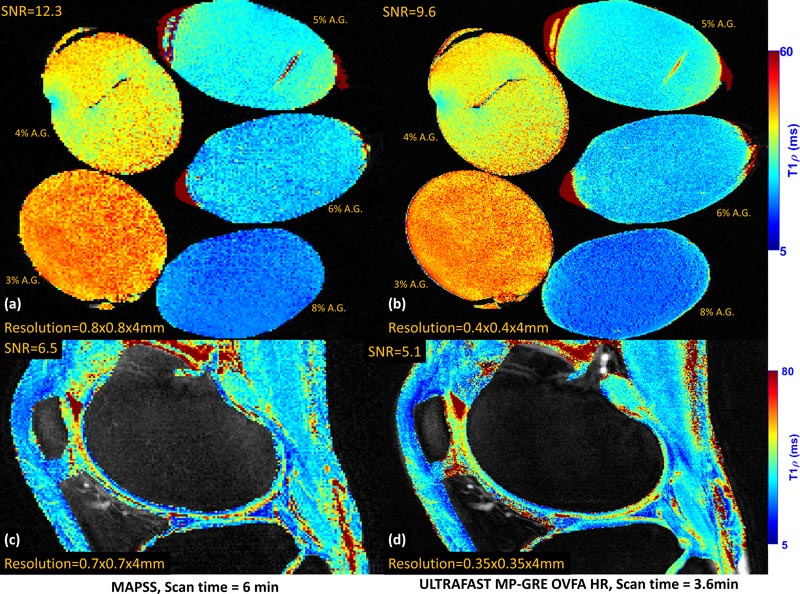
In the related preprint cited below, we find that all three sequence types have higher singal-to-noise ratio (SNR) with the data-driven optimization than without, given equal aquisition time.
In some cases, the combined benefits of higher SNR and shorter scan time are so significant as to result in image resolution four times greater than that obtained with conventionally calculated flip angles.
Related Preprint
Optimizing Variable Flip-Angles in Magnetization-Prepared Gradient Echo Sequences for Efficient 3D-T1rho Mapping
arXiv. Preprint posted online November 22, 2022. arXiv:2211.09214 [physics.med-ph]
Please cite this work if you are using MATLAB scripts for data-driven optimization of variable flip angles in MP-GRE sequences for T1ρ mapping.
References
In vivo T(1rho) mapping in cartilage using 3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (3D MAPSS)
Magn Reson Med. 2008 Feb;59(2):298-307. doi: 10.1002/mrm.21414
Three-Dimensional GRE T1ρ mapping of the brain using tailored variable flip-angle scheduling.
Magn Reson Med. 2020 Sep;84(3):1235-1249. doi: 10.1002/mrm.28198
Isotropic morphometry and multicomponent T1 ρ mapping of human knee articular cartilage in vivo at 3T.
J Magn Reson Imaging. 2018 Dec;48(6):1707-1716. doi: 10.1002/jmri.26173
Biexponential T1ρ relaxation mapping of human knee cartilage in vivo at 3 T.
NMR Biomed. 2017 Oct;30(10):10.1002/nbm.3760. doi: 10.1002/nbm.3760
Prospective Accelerated Cartesian 3D-T1rho Mapping of Knee Joint using Data-Driven Optimized Sampling Patterns and Compressed Sensing.
Proc Intl Soc Magn Reson Med. 29 (2021). p. 3310.
Get the Code
The software available on this page is provided free of charge and comes without any warranty. CAI²R and NYU Grossman School of Medicine do not take any liability for problems or damage of any kind resulting from the use of the files provided. Operation of the software is solely at the user’s own risk. The software developments provided are not medical products and must not be used for making diagnostic decisions.
The software is provided for non-commercial, academic use only. Usage or distribution of the software for commercial purpose is prohibited. All rights belong to the author (Marcelo Wust Zibetti) and NYU Grossman School of Medicine. If you use the software for academic work, please give credit to the author in publications and cite the related publications.
Contact
Questions about this resource may be directed to Marcelo Wust Zibetti, PhD.
Related Story
Marcelo Zibetti, imaging scientist at NYU Langone Health, talks about efficiency in MRI, the value of differing vantage points, and learning by thinking across disciplines.
Related Resources
MATLAB scripts for learned pulse sequence parameters of magnetization-prepared gradient echo sequences used in multi-component T2 and T1ρ mapping.
Simultaneous machine learning optimization of parallel MRI sampling pattern and variational-network image reconstruction parameters.
Machine learning optimization of k-space sampling for accelerated MRI.
An accurate, rapidly converging, low-compute algorithm for approximating solutions to the least absolute shrinkage and selection operator (LASSO) problem.