Radhika Tibrewala, MS, is a graduate student in the Biomedical Imaging and Technology PhD Training Program at NYU Grossman School of Medicine’s Vilcek Institute of Graduate Biomedical Sciences. She investigates machine learning applications to medical imaging. Her doctoral research is advised by Daniel Sodickson, MD, PhD.
Radhika is the lead author of a new data resource and accompanying preprint titled “FastMRI Prostate: A Publicly Available, Biparametric MRI Dataset to Advance Machine Learning for Prostate Cancer Imaging.” Our conversation was edited for clarity and length.
April 20, 2024 update: The FastMRI Prostate paper has been peer reviewed and published in Nature Scientific Data.
How would you describe your area of scientific research?
Before I came to NYU Langone Health, I was working primarily on musculoskeletal problems like osteoarthritis and lower-back pain, and using heavy image processing and machine learning for early diagnosis and early intervention. After coming to NYU Langone I changed tracks a little bit—I still use image processing and machine learning but I’m applying it more to accessible imaging and low-field imaging for prostate cancer detection. These are some of the themes I’ve been working on. I’m fortunate to be involved in more than one project and explore more than one technique or application.
You’ve been working with both MRI and machine learning for a while now. Which interest came first?
MRI definitely came first. I got my master’s in biomedical imaging at UCSF, where I started in 2016. Machine learning only popped up later when I was working in an osteoarthritis imaging lab—it quickly became almost essential to learn how to do machine learning for certain applications, because ML took over the imaging field in a big way.
We’re talking about 2017, 2018?
Absolutely, I started working in a the osteoarthritis lab in 2017 and was there until 2020. During those three years, I started to build my skills in MRI image processing, machine learning diagnostics, and segmentation. Then I came to NYU Grossman School of Medicine.
Your research has included machine learning applications for staging ACL injuries, estimation of muscle fat fraction, tissue segmentation and classification of lumbar spine conditions, and, more recently, segmentation from k-space with the K2S challenge. At this point, what is your perspective on the potential of deep learning for MRI?
It’s essential to have deep learning and machine learning in imaging applications, because the inherent problem with MRI is that it is slow and expensive, and at some point, we reach the limits on hardware. The solution we have to explore then is software, and given the recent advent of machine learning and deep learning, that is one huge target for a lot of people to try to make MRI faster, cheaper, better, and get it to give you the answers you need. There are so many ways deep learning is being looked at—the amount of literature on it is insane. I don’t see it plateauing anytime soon.
When we talk about machine learning applications to MRI, besides clinical or physiological areas—musculoskeletal, cardiovascular, neurological, and oncological imaging—there are also technical areas, like image reconstruction, segmentation, classification, and motion correction. Do you see any of these as having more potential for being transformed by deep learning than others?
I think they all tie into each other. Your classification algorithm will improve if your reconstruction has been improved. Your reconstruction will improve if your motion correction technique is better. They’re all connected. Ultimately, we will hopefully improve the utility of MRI and make it more accessible, and more useful not just in diagnosis but also in drug development and in understanding the origins of diseases.
Something you have just finished working on is the fastMRI prostate dataset. Why has the team decided to release this data?
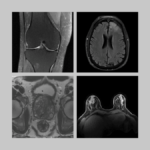
In 2018, NYU Langone released the initial fastMRI dataset, which was the first raw k-space data made publicly available, and it was kind of a big deal. It helped in exploding the field of machine learning image reconstruction and other people followed suit, making more data available. When I started working with Dan Sodickson on prostate cancer and some other projects, we said, hey, let’s keep the momentum going, let’s add more data.
The prostate dataset we’ve just released was acquired on patients who were referred for a clinical prostate MRI on suspicion of having prostate cancer, so it adds a different value. The original fastMRI was mostly a research dataset—it does have some annotations that were released later but it’s primarily for technical research on image reconstruction. With the prostate dataset, we are also adding raw k-space for an MRI diffusion sequence, which, as far as we know is a first.
So this dataset offers more than just new anatomy. The fact that it’s acquired on a clinical population means it can be used for classification studies.
That’s right, and we are actually releasing labels along with the dataset, so we’re sharing annotations for all of these cases to indicate presence or absence of prostate cancer on the MR images.
What is the size of this dataset?
It has 312 cases—a lot smaller than the knee and the brain datasets released before. The main significance of it is that it’s clinical and it has a diffusion acquisition. We think it’s worth putting out there, especially for people who are exploring different techniques for diffusion reconstruction and for diagnosis based on diffusion data.
What goes into producing such a dataset for the research community?
Because it’s a clinical population, we had to take a number of extra steps and a lot more precaution in releasing this data, so there was a massive anonymization pipeline. We also standardized all of the scans according to a vendor-neutral ISMRMRD format. We had to make sure that the images and the annotations were aligned properly and that the annotations were correct. We also had to ensure that the data were all in the same orientation in k-space. And we also built a number of reconstruction scripts that go along with the raw data processing and data preprocessing—it literally feels like we did three hundred different things to get this out there.
How long has it taken to put all this together?
I think I started working on this maybe last September, so it’s taken about six or seven months.
The related preprint has 11 coauthors. If we look at who contributed what, can it be read as a recipe of sorts? If one has, say, a team of ten people, and two of them can help with access to radiology reports, two can prep the data and the labels, two can consult on data structure, two can assist with the reconstruction code, and so on—is that a recipe for other research groups to also be able to contribute datasets like these?
I think it’s possible if you have enough expertise. There were definitely a few elements that Patricia Johnson and I were able to catch, like reading through the metadata and the headers to make sure they make sense. Somebody who hasn’t been looking at raw data or working with image reconstructions may not be able to catch that stuff. But it’s a pretty good recipe, I would say.
The ML research community always wants more data but providing annotated, curated, deidentified, raw k-space MRI data has been difficult. The preprint describes the data itself, but it doesn’t say, here’s how we did it, here’s how you put together a pipeline to produce this kind of resource.
That true. It’s usually a one-liner in the manuscript, like we took the data and deidentified it, and boom. But that does not do justice to how much work we actually had to do to get it out there.
At NYU Langone we have a lucky situation because of fastMRI, so a lot of the legal stuff is already taken care of as it was ironed out a few years ago. We still have to get the work done, which is all of the steps I mentioned: the anonymization, standardization, labeling, checking each and every patient scan, each and every slice. Working with diffusion raw data is a little bit more challenging. Diffusion has more dimensions, it’s heavier data, and the reconstruction can be variable between different groups. But because NYU Langone has set up a process with fastMRI, a lot of the sharing and hosting solutions are already in place. And in the future, a few things we’ve worked out in this release will help in the next release. So, the more we do it the easier it becomes.
This may be the only available raw k-space dataset in diffusion MR with a clinical population. Why is it important to have raw k-space data out there?
By the time you go from raw k-space to a DICOM image, the scanner itself does a lot of internal processing that you cannot undo. So the reason to have raw k-space is to have the original information acquired. It’s richer data. And now with the interest in using machine learning for speeding up reconstruction in particular, you need to have the raw data. You can’t do that from a DICOM image. So, we’re interested in seeing whether releasing diffusion raw data will encourage people to come up with more advanced, faster, better diffusion reconstruction algorithms.
Is that a big hope for the team behind this data release?
That’s one of the things—that the diffusion data sets it apart. And of course it’s a cancer dataset, so overall we’d like to see it help improve the utility of MRI for cancer diagnosis. By giving researchers raw k-space data, we’re giving them complete flexibility.
You said that your interest in MRI preceded your interest in machine learning. How did it develop?
In high school I was interested in biology and physics, and did a project on imaging—it was part of an international baccalaureate program in India, where I grew up. It was just a short overview of imaging, like, this is what an x-ray does, this is what an ultrasound does, this is what an MRI does. And I found it really cool that you can look inside the body noninvasively. Growing up, I always wanted to be in medicine, but I can’t look at blood—I instantly faint. It’s a vasovagal reaction.
Later, I went to York University in Toronto for biophysics, and I learned more about what actually happens in nuclear magnetic resonance and got to work in an fMRI lab. Afterward, I returned to India and interned at Siemens Healthiness in Mumbai. My interest in imaging just kept growing and growing, and my determination to go into a master’s program just kept increasing. To prepare for the master’s, I studied at the University of Heidelberg, remotely for a year, and two months in person. I’ve been to a lot of schools.
So you have nourished an interest in imaging over many years. What was it like for you once you were at UCSF in a program focused on imaging?
It was amazing. They have a very strong radiology department and a really good program. It was small, so we got a lot of attention and we had the opportunity to take classes throughout the year and to work through an entire thesis in the summer. My sister lives in the Bay Area, so it was also a little bit of a personal choice to move to San Francisco.
How did you make the transition from that graduate program to the PhD program at NYU Grossman School of Medicine?
I started my PhD in 2020. At that point I had worked as full-time research assistant for three years in an MRI lab. The transition was exciting—to go somewhere new, to work with new people, work on new projects. The pandemic messed a lot of stuff up. We didn’t actually get to move to New York right away—the first year of graduate school was remote. And when we did, it was coupled with so much anxiety. I basically overloaded on all of my online classes in my first year.
Tell me more about that.
I basically packed my first year with two year’s worth of coursework. Because it was mostly remote, we weren’t going to be able to start any meaningful projects that soon. And I thought, I have all this time, I don’t have to commute anywhere, I don’t have to do anything, so I loaded up on classes in order to be able to focus exclusively on research once I joined the lab.
Would you recommend that approach?
It’s nice not to have to go to class. But everyone has their own way of doing things.
What about connections with classmates?
That’s unfortunately something that was missing the first year of my doctoral study. Luckily, after I moved to New York I did make an effort to go to some of our graduate school mixers and events, and I was lucky to find a few people I have a lot of things in common with. A few of them are in imaging but most are not, so it’s actually fun to see what’s going on in their wet labs.
Students in the Vilcek Institute usually go through three lab rotations in the first year. Is that something you were able to do even though most of the work was remote?
I did a rotation with Florian Knoll while I was still remote. Then I did a rotation with Ryan Brown, and after that I joined Daniel Sodickson.
Ryan Brown leads a hardware lab. Most of your experience is in software—what was that rotation like?
At the time, Ryan was looking into doing a little bit of machine learning for an accelerometer project and needed some software development, so that was my focus.
Was it difficult for you to choose which lab you wanted to specialize in?
It was a little fortuitous. Dan Sodickson was planning a research project and I had a little bit of experience that could help, so I was in some of the meetings related to that. It turned out to be a good match and it just worked out. And of course, I wasn’t going to turn down an opportunity to work with Dan.
Now that the prostate dataset is out, what else are you working on?
I’m working on a few things. I’m actually using this dataset to develop low-field prostate cancer detection methods. This summer, I’ll be visiting the University of Napoli Federico II in Italy to work on longitudinal prostate detection and monitoring using similar data. I’m also working on an unrelated MR fingerprinting project.
You’re in the third year of your doctoral study. What are you thinking of doing after earning your PhD? Where do you think you want to be headed next?
I really enjoyed my experience interning at Genentech after my first year of PhD, so I would like to go out and use some of these skills in a biotech setting. I want to stay in imaging and do similar work but use it for drug development or clinical translation.
Related Resource
Raw k-space data and DICOM images from thousands of MRI scans of the knee, brain, and prostate, curated for machine learning research on image reconstruction.
Related Stories
The fastMRI dataset now includes curated breast MRI data to boost AI innovation in radial, dynamic contrast-enhanced, ultrafast MRI of the breast.
Patricia Johnson, who researches machine learning image reconstruction, talks about faster MRI, visual preferences, and diagnostic interchangeability.
Florian Knoll, incoming chair of imaging at University of Erlangen–Nuremberg, talks about his background, not being greedy, and why he does what he does.