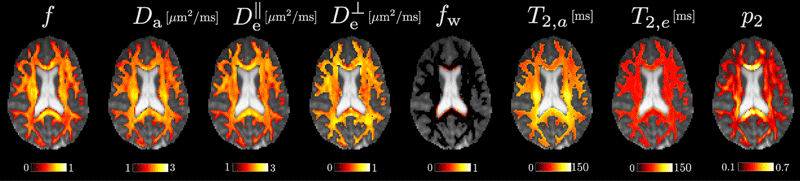
We are sharing a robust implementation of parameter estimation for the standard model of diffusion in white matter along with example datasets. The toolbox can be used to analyze diffusion MRI images and extract information about various structures present in a voxel of white matter, such as axons, extra-axonal space, and cerebrospinal fluid.
The standard model is a unifying framework of white matter diffusion models, most of which are described by the same underlying physics (NMR Biomed. 2019;32[4]:e3998. doi: 10.1002/nbm.3998). In such a framework, most diffusion models can be understood as special cases of the standard model (Neuroimage. 2018;174:518-538. doi: 10.1016/j.neuroimage.2018.03.006).

The SMI Toolbox supports:
- Linear tensor encoding data only (i.e. single diffusion encoding) and equal echo time (TE) in all diffusion-weighted images.
- Multiple tensor encodings and equal TE in all diffusion-weighted images.
- Multiple tensor encodings and differing TE between all diffusion-weighted images (when multiple TE data are used, the SMI Toolbox returns axonal and extra-axonal T2 estimates).
For best results, we suggest preprocessing the diffusion-weighted data with the DESIGNER pipeline, which outputs a noise map that can be used as an input for the SMI Toolbox.
Related Publications
Reproducibility of the Standard Model of diffusion in white matter on clinical MRI systems.
Neuroimage. 2022 May 8:119290. doi: 10.1016/j.neuroimage.2022.119290
Rotationally-invariant mapping of scalar and orientational metrics of neuronal microstructure with diffusion MRI.
Neuroimage. 2018;174:518-538. doi: 10.1016/j.neuroimage.2018.03.006
Please cite these works if you are using the SMI Toolbox in your research.
References and Further Reading
Open this card for a handpicked list of publications grouped by standard-model features: the stick compartment model, constrained and unconstrained model versions, model degeneracies, parameter estimation with machine learning, sensitivity and specificity, model validation, reproducibility, and experimental design.
References and Further Reading
Open this card for a handpicked list of publications grouped by standard-model features: the stick compartment model, constrained and unconstrained model versions, model degeneracies, parameter estimation with machine learning, sensitivity and specificity, model validation, reproducibility, and experimental design.
Stick Compartment Model of Axons
On the nature of the NAA diffusion attenuated MR signal in the central nervous system.
Magn Reson Med. 2004 Nov;52(5):1052-9. doi: 10.1002/mrm.20260
Modeling dendrite density from magnetic resonance diffusion measurements.
Neuroimage. 2007 Feb 15;34(4):1473-86. doi: 10.1016/j.neuroimage.2006.10.037
Constrained Versions of the Standard Model
White matter characterization with diffusional kurtosis imaging.
Neuroimage. 2011 Sep 1;58(1):177-88. doi: 10.1016/j.neuroimage.2011.06.006
NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain.
Neuroimage. 2012 Jul 16;61(4):1000-16. doi: 10.1016/j.neuroimage.2012.03.072
Multi-compartment microscopic diffusion imaging.
Neuroimage. 2016 Oct 1;139:346-359. doi: 10.1016/j.neuroimage.2016.06.002
Unconstrained Standard Model Formulation
Rotationally-invariant mapping of scalar and orientational metrics of neuronal microstructure with diffusion
MRI.
Neuroimage. 2018 Jul 1;174:518-538. doi: 10.1016/j.neuroimage.2018.03.006
Quantifying brain microstructure with diffusion MRI: Theory and parameter estimation.
NMR Biomed. 2019 Apr;32(4):e3998. doi: 10.1002/nbm.3998
Standard Model Degeneracies
Degeneracy in model parameter estimation for multi-compartmental diffusion in neuronal tissue.
NMR Biomed. 2016 Jan;29(1):33-47. doi: 10.1002/nbm.3450
Resolving degeneracy in diffusion MRI biophysical model parameter estimation using double diffusion encoding.
Magn Reson Med. 2019 Jul;82(1):395-410. doi: 10.1002/mrm.27714
A unique analytical solution of the white matter standard model using linear and planar encodings.
Magn Reson Med. 019 Jun;81(6):3819-3825. doi: 10.1002/mrm.27685
Parameter Estimation with Machine Learning
Disentangling micro from mesostructure by diffusion MRI: A Bayesian approach.
Neuroimage. 2017 Feb 15;147:964-975. doi: 10.1016/j.neuroimage.2016.09.058
Reproducibility of the Standard Model of diffusion in white matter on clinical MRI systems.
Neuroimage. 2022 Aug 15;257:119290. doi: 10.1016/j.neuroimage.2022.119290
How do we know we measure tissue parameters, not the prior?
Proc Intl Soc Magn Reson Med. 29 (2021). p 0397
Sensitivity Specificity Matrix for Assessing Parameter Estimation
Mapping tissue microstructure of brain white matter in vivo in health and disease using diffusion MRI.
Imaging Neurosci (Camb). 2024; 2 1-17. doi: 10.1162/imag_a_00102
Model Validation
Volume electron microscopy in injured rat brain validates white matter microstructure metrics from diffusion
MRI.
Imaging Neurosci (Camb). 2024; 2 1-20. doi: 10.1162/imag_a_00212
Reproducibility
Reproducibility of the Standard Model of diffusion in white matter on clinical MRI systems.
Neuroimage. 2022 Aug 15;257:119290. doi: 10.1016/j.neuroimage.2022.119290
Rotationally-invariant mapping of scalar and orientational metrics of neuronal microstructure with diffusion
MRI.
Neuroimage. 2018 Jul 1;174:518-538. doi: 10.1016/j.neuroimage.2018.03.006
Optimal Experimental Design
Optimal experimental design for biophysical modelling in multidimensional diffusion MRI.
arXiv. Preprint posted online July 13, 2019. doi: 10.48550/arXiv.1907.06139
Towards unconstrained compartment modeling in white matter using diffusion-relaxation MRI with tensor-valued
diffusion encoding.
Magn Reson Med. 2020 Sep;84(3):1605-1623. doi: 10.1002/mrm.28216
Reproducibility of the Standard Model of diffusion in white matter on clinical MRI systems.
Neuroimage. 2022 May 8:119290. doi: 10.1016/j.neuroimage.2022.119290
Rotationally-invariant mapping of scalar and orientational metrics of neuronal microstructure with diffusion
MRI.
Neuroimage. 2018;174:518-538. doi: 10.1016/j.neuroimage.2018.03.006
Patents
System, method and computer-accessible medium for determining brain microstructure parameters from diffusion
magnetic resonance imaging signal’s rotational invariants.
US Patent 10,360,472. July 23, 2019.
System, method and computer-accessible medium for determining rotational
invariants of cumulant expansion from one or more acquisitions which can be minimal.
International patent application PCT/US2023/019,622. April 24, 2023.
Get the Code
This resource is maintained on Github as Standard Model Imaging (SMI) Toolbox.
Example Data
Below, we provide three example datasets for easy testing of the SMI toolbox. All data were preprocessed with DESIGNER.
dataset_1contains a 2-shell diffusion MRI protocol similar to the one in the UK biobank.dataset_2contains multiple b-value b-tensor shape combinations (all with the same echo time), similar to the one we proposed in the related publication cited above.dataset_3contains multiple combinations of b-values, b-tensor shapes, and echo times.
Note that all three datasets are provided in nifti format. We used Jimmy Shen’s Tools for NIfTI and ANALYZE image toolbox to handle the data in MATLAB but users may turn to other tools of their choice and modify the lines in the example code.
Get the Data
The data available on this page are provided free of charge and comes without any warranty. CAI²R and NYU Grossman School of Medicine do not take any liability for problems or damage of any kind resulting from the use of the files provided. Operation of the software is solely at the user’s own risk. The software developments provided are not medical products and must not be used for making diagnostic decisions.
The software is provided for non-commercial, academic use only. Usage or distribution of the software for commercial purpose is prohibited. All rights belong to the author (Santiago Coelho) and NYU Grossman School of Medicine. If you use the software for academic work, please give credit to the author in publications and cite the related publications.
Contact
Questions about this resource may be directed to Santiago Coelho, PhD, or raised as issues on Github.
Related Story
Santiago Coelho, postdoctoral fellow who develops diffusion MRI methods for brain imaging, talks about modeling tissue properties, entering the field by chance, and what he proposes to do next.

